Hi everyone, I'm Eric, a data scientist here at Dataiku. I'm going to be doing a series of blog posts about using Dataiku DSS in biotech. So stay tuned and check back for more biotech-specific posts!
If you work in biotech or bioinformatics, you have probably come across the dreaded Variant Call Format. I spent many sleepless nights during my Ph.D. trying to parse these files. Why does the same file have semicolons, tabs and commas? Why key-value pairs come in non-standard formats? Why? WHY?? Only the bioinformatics gods (a.k.a. the researchers at 1000 genomes) know.
The goal of this blog post is to create a VCF parser in Dataiku DSS. We're going to do this in less than 10 minutes — for real. The parser is going to be fast and based on our streaming architecture, which means you don't have to worry about those pesky memory issues. And — wait for it — it's going let you store the output where ever you want: your filesystem, SQL systems, NoSQL systems, or the cloud.
You ready? Me too.
Loading the Data
I'm going to build this with chromosome Y from the 1000 genomes project. Chromosome Y is the smallest (only 20 genes!), so it's the easiest for demo purposes.
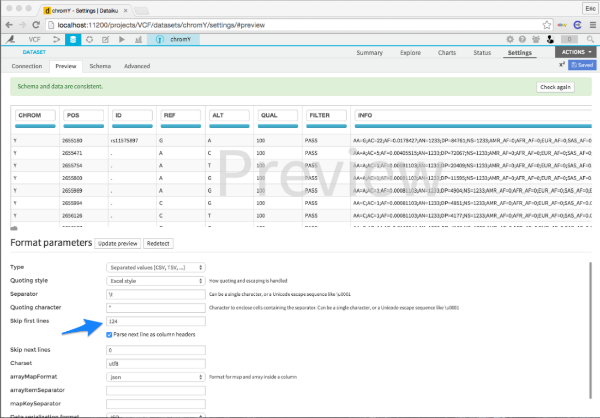
To upload the data, I just select Dataset > Upload your file, and then I clicked and dragged the VCF into the interface. I then told Dataiku DSS to skip all the comment lines (124 in this case) and that's it! I clicked create and I had my VCF file in my project.
Parsing the Info Field
Hadley Wickham, the R guru and all around data science rockstar, has an excellent paper on tidy vs. messy data. If you haven't read it, you should head over right now. It'll change the way you analyze data.
The info column in a VCF file is a prototypical example of messy data. The column has multiple entries separated by a semi-colon. Those entries can either be key-value pairs separated by an equals sign or just a key without a value. Hadley Wickham does not approve.
We're going to use Dataiku DSS to clean this data. First, I'm going to make a unique ID for each row. Then, I'm going to split the entries in info by the semi-colon, then fold, then split on an equals sign, then pivot. Sound complicated? It is. But! Dataiku DSS gives you visual feedback every step of the way so you know what you're doing.
I made a video showing the whole process. It takes just over four minutes to do the whole thing. Check it out!
Folding Genotypes
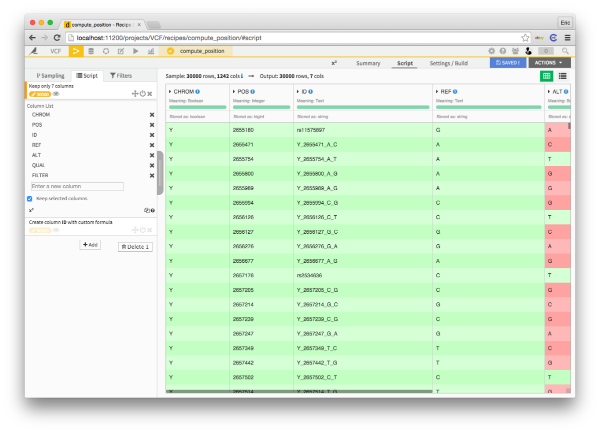
Next up, we have the actual genotypes. The VCF file represents each individual as a column and each position as a row. This format is fine, but I prefer to have my data in the long-and-skinny format, rather than the short-and-fat format. Group-by operations are more flexible with long-and-skinny data, and everyone loves group-bys. To transform my data from short-and-fat to long-and-skinny, I'm going to use a fold processor.
Selecting Other Columns
Finally, we need the columns with the position, reference allele, and alternative allele for each variant. I'm going to make just a simple recipe to select those columns and put them into another dataset.
Deployment!
Great, we've got our parser working. We're separating the mess of a VCF file into three separate datasets: info about each variant, the position of each variant and, finally, the genotype for each patient x variant pair.
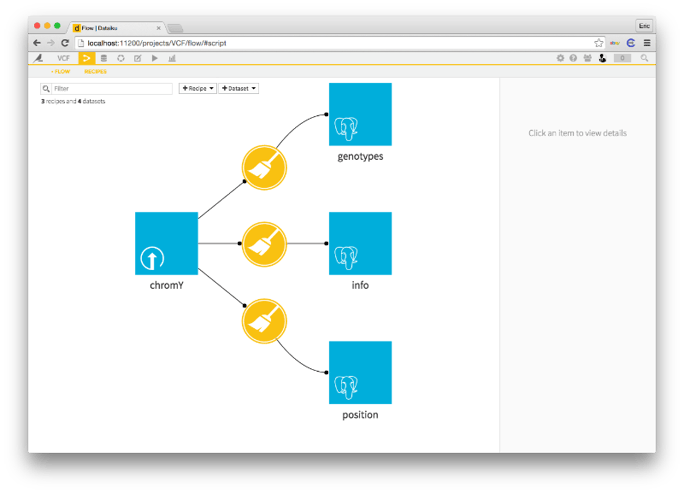
You may notice the little elephant heads in the bottom left of each of the final three datasets. That's because they're in PostgreSQL! You can choose where you want your data to go after the parser. You can put the result back onto your normal filesystem, into a SQL system, into a NoSQL system or HDFS. The choice is yours and yours alone.




